Theme 3
Enabling Algorithms
Enabling Algorithms (EAs) are general and efficient methods to do (mostly) numerical and graphical analyses of real and synthetic data; such algorithms are needed across all areas of ACEMS research. For example, we may wish to fit a time series model to hydrology data and use it for prediction. The EA would estimate the model, usually expressed in equation form, evaluate how well it fitted, and then develop prediction methods. As researchers refine their understanding of a subject and with greater access to data, current EAs need to be refined and more powerful ones created.
Throughout its existence, and in particular in 2021, ACEMS researchers have worked on a range of computational enabling algorithms, including: developing new and enhanced algorithms to solve a variety of methodological and applied problems; showing both theoretically and empirically that the algorithms work as claimed; and clearly describing and documenting the algorithms to make it easier to understand and code them. During 2021, the EA researchers made significant advances in key areas that are used to analyse and solve questions in a wide variety of fields, including numerical linear algebra, advanced particle filter methods for estimating complex time series models, approximate methods for Bayesian analysis that can handle complex models and large data sets, with applications to computational intelligence, system reliability, agriculture, sports and epidemic modelling.
People
Kevin Burrage, Tim Garoni, Rob Hyndman, Robert Kohn*, Dirk Kroese*, Kerrie Mengersen, Scott Sisson, Kate Smith-Miles, Ian Turner*, Matt Wand (*Theme Leaders)
Azam Asanjarani, Peter Ballard, Nigel Bean, Boris Beranger, Andrew Black, Pamela Burrage, Julian Caley, Jess Cameron, Andrea Collevecchio, Dianne Cook, Tiangang Cui, Simon Denman, Chris Drovandi, Megan Farquhar, Troy Farrell, Denzil G. Fiebig, David Frazier, Clara Grazian, Jens C Grimm, David Gunawan, Shovanur Haque, Sophie Hautphenne, Markus Hegland, Kate Helmstedt, Jacinta Holloway-Brown, Wang Jin, Sevvandi Kandanaarachchi, Jonathan Keith, Bonsoo Koo, Brodie Lawson, Catherine Leigh, Jess Liebig, Benoit Liquet, Gael Martin, James McGree, Ross McVinish, Pablo Montero Manso, Mario Andres Munoz Acosta, Giang Nguyen, Steven Psaltis, Rachael Quill, Matias Quiroz, Ben Rohrlach, Fred Roosta, Joshua Ross, Robert Salomone, Leah South, Thomas Taimre, Priyanga Dilini Talagala, Thiyanga Talagala, Emi Tanaka, Duncan Taylor, Nick Tierney, Minh Ngoc Tran, Radislav Vaisman, Mattias Villani, Paul Wu, Nan Ye
Elma Akand, Jessica Benitez Mendieta, Xuhui Fan, Puwasala Gamakumara, Lachlan Gibson, Marcus Hoerger, Stuart Lee, Sarat Babu Moka, Matthieu Simon, Chris van der Heide, David Warne, Eric Zhou
Juan Muriel, Mitchell O’Hara-Wild
Igor Balnozan, Edward Barker, Mark Baxendale, Joshua Bon, Sophie Chen, Rixon Crane, Laurence Davies, Mohammad Davoudabadi, Dilishiya De Silva, Vektor Dewanto, Zhe Ding, Ethan Goan, Thomas Goodwin, Sayani Gupta, Adam Hamilton, Virginia He, Wathsala Karunarathne, Shamika Prasadini Kekulthotuwage Don, Jun Ju, Aline Kunnel, Yeming Lei, Angus Lewis, Dan Li, Yang Liu, Amalan Mahendran, David Maine, Abraj Mohomed Haseem Mohomed Amsar, Ryan Moseni, Aby Nasrawi, Aaron Snoswell, Shian Su, James Walker, Riley Whebell, John Worrall, Yu Yang, Alice Yao
Zeke Ahern, Hugh Andersen, Sarah Belet, Imke Botha, Adam Bretherton, Valentina di Marco, Nicholas Johnson, Ryan Kelly, Yang Liu, Dylan Morris, Jon Peppinck
Mitchel O'Sullivan, Jacinta Roberts
Key Achievements
In 2021, ACEMS researchers continued developing and analysing novel methodologies and improving existing methods to solve a range of problems. Researchers in this theme made significant advances in key algorithms that are used to analyse and solve questions in a wide variety of fields, including numerical linear algebra, computational intelligence, system reliability, agriculture, sports and epidemic modelling. Some highlights are outlined below.
CI Matt Wand published an R package named densEstBayes on the Comprehensive R Archive Network (https://www.R-project.org/). This software module facilitates the use of several Bayesian inference engine approaches, including some developed within Wand's group, for obtaining Bayesian density estimates.
ACEMS members continued to share their latest research throughout 2021 in various ways. AI Ross McVinish published, jointly with former ACEMS PhD student Liam Hodgkinson (now at UC Berkeley), a paper in the top-ranking Annals of Applied Probability, on approximate simulation of finite long-range spin systems. RF Chris van der Heide, AI Fred Roosta and CI Dirk Kroese presented and published their paper on Shadow Manifold Hamiltonian Monte Carlo in the prestigious International Conference on Artificial Intelligence and Statistics. ACEMS PhD students also presented virtually at MODSIM2021, with Alice Yao giving the talk "Estimating Tail Probabilities of Random Sums of Scale Mixture of Phase-type Random Variables", while Yeming Lei presented a new Bayesian method for fishery stock assessment. RF Lachlan Gibson’s presentation titled A Novel Implementation of Q-Learning for the Whittle Index, won the Best Community-voted Presentation Award at the EAI Valuetools 2021 conference. Meanwhile, AI Thomas Taimre published a Springer monograph, jointly with former ACEMS PhD Student Patrick Laub and Young Lee, on The Elements of Hawkes Processes.
AI Fred Roosta and his students developed new and improved algorithms for novel second-order optimization methods for neural networks. The findings were published in the prestigious Conference on Uncertainty in Artificial Intelligence (UAI2021). AI Radislav (Slava) Vaisman developed and analyzed several new Monte Carlo algorithms for efficient network reliability estimation and simulation.
The ACEMS research team consisting of AI Steven Psaltis and CI Ian Turner worked closely with other collaborators and the Centre’s Industry Affiliate Member Queensland Department of Agriculture and Fisheries, Forest Products Innovation Team (QDAF) to assist the forest sector with the optimisation of the wood value chain through enriching the timber industry’s capability in precision forestry. This fundamental research is important because the success of the sector is nowadays more and more dependent on value optimisation of their resource, rather than the traditional volume optimisation. The team used a combination of advanced data analytics, mathematical modelling, and simulation to help the Queensland forestry industry better predict the value of their Southern Pine plantation resource. Their findings informed industry on the best time to harvest, as well as the optimal sawing pattern and drying schedule to apply to maximise profits, thus enhancing management decision-making.
Read more about this exciting project here.
During 2021, the ACEMS research team consisting of AI Steven Psaltis and CI Ian Turner worked closely with other collaborators and the Centre’s long-term Industry Affiliate Member, the Queensland Department of Agriculture and Fisheries (QDAF) (LINK TO PAGE) Forest Products Innovation Team to assist the forest sector with the optimisation of the wood value chain through enriching the timber industry’s capability in precision forestry. This fundamental research is important because the success of the sector is nowadays more and more dependent on value optimisation of their resource, rather than the traditional volume optimisation. The team used a combination of advanced data analytics, mathematical modelling, and simulation to help the Queensland forestry industry better predict the value of their Southern Pine plantation resource. Their findings informed industry on the best time to harvest, as well as the optimal sawing pattern and drying schedule to apply to maximise profits, thus enhancing management decision-making.
An important observation is that careful analysis of log stiffness properties can lead to not only an improved financial return for the forest sector, but also a reduction in overall wood wastage. Sawn timber is the main product manufactured from softwood resources and board value depends strongly on its mechanical performances. Wood stiffness is thought to be a key factor driving forest value, and the non-structural and structural grades of boards are specified based on product stiffness quantified by the modulus of elasticity (MOE). The MOE dictates the mechanical grade of the board, and the market value of the board is directly linked to its grade. The market value of a board is based on individual grading performance, with structural grade boards worth more than four times the value of non-structural boards.
A significant outcome of the work was the use of increment cores to enhance predictive capabilities of the MOE of sawn boards (see Figure 1).
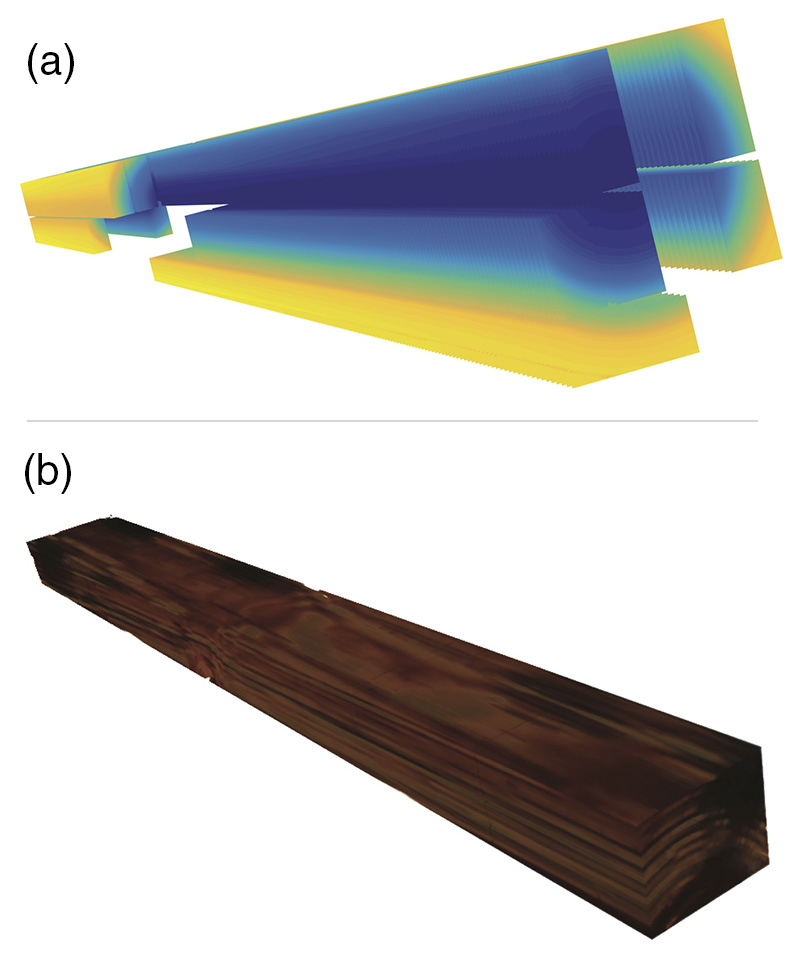
Figure 1. (a) Predicted board MOE based on increment core data. (b) Reconstructed board based on image data from peeled veneer.
This work resulted in two journal articles published in the Annals of Forest Science and one in Computers and Electronics in Agriculture.
The Forest and Wood Products Australia project conducted in association with several university and industry partners saw mathematical modelling techniques applied to improve returns from Southern Pine plantations in Southeast Queensland and Northern NSW through innovative resource characterization modelling. The team found that sigmoid curves developed to fit the measured data captured the variation in wood properties from the pith to the bark quite well and provided valuable information on product performance (see Figure 2).
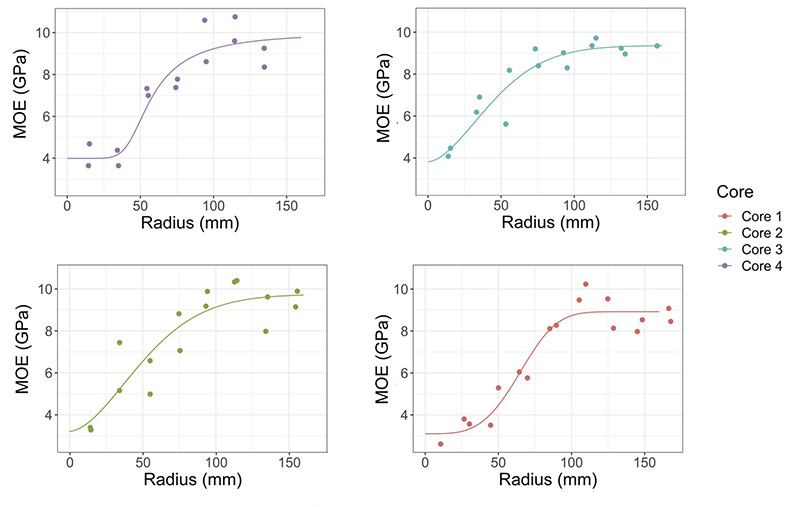
Figure 2. (a) Predicted board MOE based on increment core data. (b) Reconstructed board based on image data from peeled veneer.
The cambial age versus ultrasound MOE allowed wood practitioners to predict the age when a stand will produce high quality wood, which improved their knowledge of how to value pine plantations.
Steve and Ian also worked on a related project with QDAF to model the impact of continuous drying on key production and performance criteria of engineered wood structural elements. Their numerical simulations showed excellent agreement with the experimental drying data, and the research informed industry on the suitability of this drying technology for their current timber processing operations. This work has the potential to lead to larger-scale collaborative projects.
This work also led to a scoping study with Hyne Timber through the Building 4.0 CRC Ltd entitled ‘Data analytics for structural fibre resources optimisation’, and another project with QDAF on modelling moisture ingress into engineered wood products. This work will be of critical importance for understanding the performance of products such as cross-laminated timber (CLT) and laminated veneer lumber (LVL) used in mid-rise timber building construction, as they are exposed to climatic elements.
Related publications
[1] Kumar, C., Psaltis, S., Bailleres, H. et al. Accurate estimation of log MOE from non-destructive standing tree measurements. Annals of Forest Science 78, 8 (2021).
[2] Psaltis, S., Kumar, C., Turner, I. et al. A new approach for predicting board MOE from increment cores. Annals of Forest Science 78(3), 78 (2021).
[3] Psaltis, S., Turner, I., Carr, E. J., Farrell, T., Hopewell, G., Baillères, H. Three-dimensional virtual reconstruction of timber billets from rotary peeling. Computers and Electronics in Agriculture 152, (2018).
[4] Baillères H, Lee D, Kumar C, Psaltis S, Hopewell G, Brancheriau L Improving returns from southern pine plantations through innovative resource characterisation. Tech. report, Forest & Wood Products Australia, Project number: PNC361-1415 (2019).
AI Chris Drovandi continued research on synthetic likelihood that was originally developed during his DECRA Fellowship (2016-2019), which has proven to be a useful method for likelihood-free inference. This has been a cross-node collaborative effort between ACEMS nodes at QUT, Monash and UNSW. With AI David Frazier, the team developed a new synthetic likelihood method that is robust to model misspecification and is able to identify which summary statistics the model is not compatible with, which was published in the Journal of Computational and Graphical Statistics (JCGS). Furthermore, with AI Frazier as well as CI Scott Sisson, CI Ian Turner and ACEMS MPhil student Jacob Priddle, the team greatly improved the computational efficiency of synthetic likelihood whilst sacrificing little on accuracy using whitening transformations (to appear in JCGS). With ACEMS PhD students Leah South and Ziwen An, their paper on an R package for Bayesian synthetic likelihood methods published in the Journal of Statistical Software. This will help to improve the accessibility of synthetic likelihood methods.
As part of an ARC Discovery Project, AI Chris Drovandi is working with ACEMS and external collaborators on developing new sequential Monte Carlo (SMC) methods and applying them in challenging applications. SMC and related particle filters are useful for Bayesian inference in complex models. With ACEMS PhD student Josh Bon (who completed in 2021) and external collaborator Prof Anthony Lee (Bristol University), the team developed new SMC methods that optimally exploit a surrogate model in order to speed-up calculations. This work was published in Statistics and Computing. Together with ACEMS PhD student Imke Botha and CI Robert Kohn, they developed new particle filtering methods for stochastic differential equation mixed effect models (published in Bayesian Analysis). Successful applications of SMC methods have been in finance models (with ACEMS PhD student Dan Li), soil carbon sequestration (with ACEMS PhD student Mohammad Javad Davoudabadi, AI Gentry White and external collaborator Dr Dan Pagendam), history matching (with Dr Pagendam and external collaborator A/Prof David Nott) and electrocardiography (with AI Brodie Lawson, CI Kevin Burrage, and external collaborators Dr Julia Camps and Prof Blanca Rodriguez).
Bayesian Networks (BNs) have been widely applied across a wide range of domains including environmental, medical, information technology domains, and as expert systems. However, although the method represents relationships between factors in a network via conditional probabilities, the parameters of the model are inherently deterministic. ACEMS AI Paul Wu and CI Kerrie Mengersen worked on methods for full Bayesian inference in a BN with potentially multiple observations in an arbitrary network of factors. Early results show promise for a Markov chain Monte Carlo (MCMC) approach assuming independence between Markov families. This was presented as part of an invited talk entitled “Bayesian Network Inference With Uncertain Evidence and Parameters” at the BISP-12 conference. Ongoing work includes: (i) development of a clustering algorithm to enable faster and possibly parallel computation, to support Bayesian inference in BNs, and (ii) Bayesian inference without restrictive independence assumptions. The work is conducted as part of an ongoing international collaboration with former ACEMS visitor Fabrizio Ruggeri (CNR-IMATI, Italy).
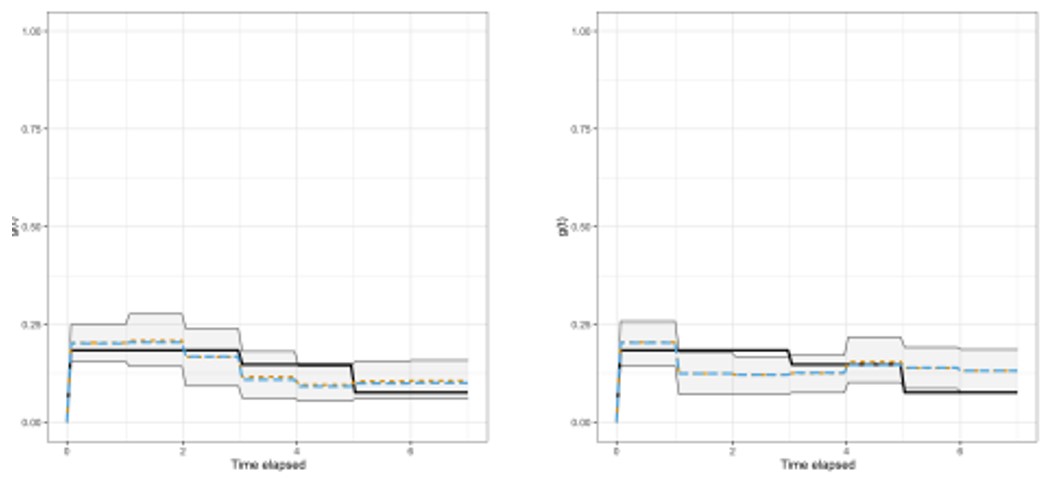
ACEMS AI Edgar Santos Fernandez led an international team of researchers in the development and application of new methods for deriving the intrinsic dimension (ID) of a dataset. The approach involves fitting a mixture of Pareto distributions to the ratio of the first and second order distances between observations, thereby allowing for multiple ID manifolds reflecting heterogeneity in the data. Truncated and repulsive priors were employed to derive new computational methods for ID analysis and clustering in a Bayesian framework. The following figure shows the ID trajectory during shots for goal in a basketball game. The solid line and coloured bands represent the median ID and 95% credible intervals, respectively, in short (red), mid-range (blue) and 3-point (green) plays, based on short and long durations of possession (left and right columns, respectively).
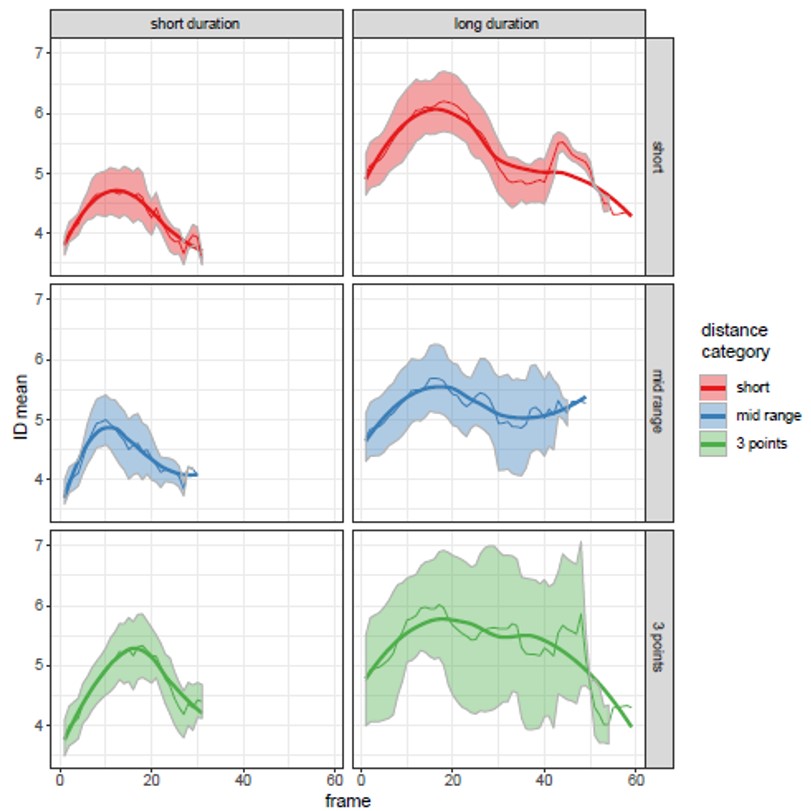
This figure shows the ID trajectory during shots for goal in a basketball game. The solid line and coloured bands represent the median ID and 95% credible intervals, respectively, in short (red), mid-range (blue) and 3-point (green) plays, based on short and long durations of possession (left and right columns, respectively).
ACEMS PhD student Raiha Browning pursued research into Bayesian nonparametric models and algorithms for discrete-time Hawkes processes. These are a form of self-exciting process, first introduced as a continuous-time point process to describe phenomena where past events increase the probability of future events occurring. Most standard models of Hawkes processes rely on a parametric form for the function describing the influence of past events, referred to as the triggering kernel. This is likely to be insufficient to capture the true excitation pattern, particularly for complex data. By utilising trans-dimensional MCMC inference techniques, Raiha developed a new model for the triggering kernel that can take the form of any step function since the location and heights of each step are unknown. This affords much more flexibility than a parametric form. A key finding is how the balance between prior informativeness and the informativeness of the data affects inference in this high-dimensional parameter space. At least one of these is required to produce reliable inference. For higher-dimensional problems, this is of particular importance. The proposed model was also applied to real-world data on COVID-19 and terrorism. This research was undertaken with international collaborator and co-supervisor Professor Judith Rousseau at Oxford University. The work is currently being extended to include a new reversible-jump Markov chain Monte Carlo algorithm for trans-dimensional estimation of the triggering function.
A histogram representation of a simulated triggering kernel over elapsed time stamps of 100 days (left) and 200 days (right). The true step function (solid line) is compared with the posterior mean and median (dashed lines) and an 80% posterior interval (grey ribbon) under an informative prior setting.
ACEMS PhD student Conor Hassan and ACEMS AI Rob Salomone have been developing Bayesian deep generative neural network algorithms to generate synthetic tabular data. These new methods will enable creation of probabilistic models, ease constraints around sharing sensitive and private data and facilitate more open access to data. Conor presented at a workshop on synthetic data that was held with the Australian Data Science Network (ADSN) – of which ACEMS is a partner and key contributor – which was attended by researchers and industry members across Australia. Industry partners in attendance included Cancer Council Queensland (CCQ), ABS, Integrity Systems Company (ISC), Sax Institute, and Services Australia. A review paper and software package are in progress.
Seleted publications produced by ACEMS members during 2021
Book
- Laub, P., Lee, Y., & Taimre, T. (2021) The Elements of Hawkes Processes. Cham: Springer International Publishing. https://doi.org/10.1007/978-3-030-84639-8.
Journal publications
- Hirsch, C., Moka, S.B., Taimre, T., & D.P. Kroese. (2021) Rare Events in Random Geometric Graphs. Methodology and Computing in Applied Probability. https://doi.org/10.1007/s11009-021-09857-7.
- Kumar, C., Psaltis, S. T. P., Bailleres, H., Turner, I., Brancheriau, L., Hopewell, G., Carr, E. J., Farrell, T., & Lee, D. J. (2021). Accurate estimation of log MOE from non-destructive standing tree measurements. Annals Of Forest Science, 78(1), 8. https://doi.org/10.1007/s13595-021-01031-w
- McVinish, R., & Hodgkinson, L. (2021). Fast approximate simulation of finite long-range spin systems. Annals of Applied Probability, 31(3), 1443 - 1473. https://doi.org/10.1214/20-AAP1624
- Psaltis, S., Kumar, C., Turner, I., Carr, E. J., Farrell, T., Brancheriau, L., Bailléres, H., & Lee, D. J. (2021). A new approach for predicting board MOE from increment cores. Annals of Forest Science, 78(3), 78. https://doi.org/10.1007/s13595-021-01093-w
- Vaisman, R. (2022) Finding minimum label spanning trees using cross-entropy method, Networks, 79(2), 220– 235. https://doi.org/10.1002/net.22057
- Vaisman, R., & Sun, Y. (2021). Reliability and importance measure analysis of networks with shared risk link groups. Reliability Engineering & System Safety, 211, 107578. https://doi.org/10.1016/j.ress.2021.107578
Conference publications
- Gibson, L. J., Jacko, P., Nazarathy, Y., Zhao, Q., & Xia, L. (2021). A Novel Implementation of Q-Learning for the Whittle Index. 14th EAI International Conference, VALUETOOLS 2021 (Vol. 404, pp. 154 - 170). Virtual: Springer. https://doi.org/10.1007/978-3-030-92511-6_10.
- Ju, J., Kurniawati, H., Kroese, D., & Ye, N. (2021) MOOR: Model-based Offline Reinforcement Learning for Sustainable Fishery Management. MODSIM2021, 24th International Congress on Modelling and Simulation. Modelling and Simulation Society of Australia and New Zealand, (pp. 771–777). Sydney: MSSANZ. https://doi.org/10.36334/modsim.2021.m2.ju.
- Lei, Y., Zhou, S., & Ye, N. (2021) Prior versus Data: A New Bayesian Method for Fishery Stock Assessment. MODSIM2021, 24th International Congress on Modelling and Simulation. Modelling and Simulation Society of Australia and New Zealand, (pp. 43–49). Sydney: MSSANZ. https://doi.org/10.36334/modsim.2021.A1.lei.
Project reports
- Filar, J. A., Courtney, A. J., Gibson, L. J., Jemison, R., Leahy, S. M., Lei, Y., Mendiolar, M., Mitchell, J., Robson, B., Steinberg, C., Williams, S. M., Yang, W.-H., Ye, N. (2021) Modelling environmental changes and effects on wild-caught species in Queensland: environmental drivers: final report. FRDC report.
ACEMS researchers worked on a large-scale collaborative project with DAF and University of the Sunshine Coast to develop a virtual sawing approach that can predict the modulus of elasticity of individual boards that can be obtained from a forest without cutting the trees. This research can significantly impact a mill’s profitability, allowing them to match timber received from growers to the end product specification requirements, and reduce the risk of sawing logs from plots not producing suitable products."
Dr Chandan Kumar, Queensland Department of Agriculture and Fisheries.